4.6. Exercises#
The exercises below are intended for work in VS Code, not within the book. You can download each exercise file (.py) and open it in your VS Code. A shortcut way of doing this is dragging and dropping files from the little download icon in the browser into your IDE. If the file you download from the book is .zip, one of the files contained within it is either a supplementary image or an answer-checker .py file.
Each exercise carries difficulty level indication:
[*] Easy
[**] Moderate
[***] Advanced
The highest difficulty level is meant for students who wish to challenge themselves and/or have previous experience in programming.
Exercise 4.8 ([*] Numerics)
For this exercise, you can use the following documentation.
An important introduction
In Python, you can print the value of variables, which is an easy way to see whether your code was effective. You can do so using the print() function; put the statement or variable you want to print between the brackets.
Run the following cell to print the sentence “Hello world!”
print("Hello world!")
Numeric types
A numeric variable, or quantitative variable, is simply a number. There are integers, which are whole numbers; floats, which are essentially decimals; and complex numbers. Note that in Python, the complex number \(i\) is represented by a \(j\)!
First, define an integer:
integer1 = # Define an integer here
print("The type of integer1 is ", type(integer1)) # The print command allows you to see what the type is.
Now define a float:
float1 = # Define a float here
print("The type of float1 is ", type(float1))
Add your integer and your float together. You can do so by using the + operator. Check what is the type of the sum using print() and type() as in the exercise above.
sum_integer_and_float =
# Write code to print the type
As you can see from the examples above, you do not have to define the type of the variable yourself; Python will do this for you. This is very convenient, since you can simply define variables and do math without having to worry about the type.
Define a complex variable:
complex1 = # Define a complex number here
print("The type of complex1 is ", type(complex1))
Add your float and your complex number together. What happens? Why? Add your answer here.
sum_float_and_complex =
Exercise 4.9 ([*] Numeric operators)
You can use the following documentation for this exercise.
Figure out what the following operators do using the variables a and b:
, - , *, /, %, //
a = 9
b = 2
# Example for +
c = a + b
print(c)
# Your code here
Exercise 4.10 ([*] Numerics type conversion)
For more information on this exercise, you can use this link.
Python infers the type of the variable based on context. In this exercise, we will explore three ways in which you can change the type of a variable by coding it in a specific way. We will do so by changing an integer into a float.
initial_integer = 5
print(type(initial_integer))
Redefining a variable
When you redefine a variable (using =), Python immediately changes the type of the variable to match the definition. Redefine the variable initial_integer to make it a float (hint: remember the definition of a float?).
initial_integer = # Put your code here
print(type(initial_integer))
As you can see, as we redefined the variable into one containing a decimal point, Python now recognizes it as a float. For the next exercise, we need to go back to our initial integer; we do so by running the following code.
initial_integer = 5
print(type(initial_integer))
Using Python’s built-in function
To make a variable of a specific type, we can use Python’s built-in type converters. To change a variable into a float, we use float(). To change it into an integer, we use int(). Convert the variable initial_integer to a float using the built-in converter.
initial_integer = # Put your code here
print(type(initial_integer))
Note that this will only work if Python can reasonably recognize the type you’re converting to in the variable. For example, when we try to covert a complex number into a float, we run into a problem:
complex_number = 5 + 1j
converted = float(complex_number)
For the next exercise, we again have to redefine the original integer so we can explore the last method to convert it into a float. Run the following code:
initial_integer = 5
print(type(initial_integer))
Using calculations
As you perform calculations, Python will infer the new type based on the operations that you use. This is of course very useful, as there are many examples of going from an integer to a float by performing certain operations.
Convert the variable initial_integer to a float by using numeric operators.
initial_integer = # Put your code here
print(type(initial_integer))
Exercise 4.11 ([*] Boolean variables)
A Boolean variable can take two values: True or False. You can use it for applying logic in Python, which you will encounter later in the book. As you saw in the last exercise, you can convert variables to another type. By converting a variable to a Boolean object, you can check the “truth value” that Python assigns to the variable.
Check the truth value of the following variable values by using bool() and print().
a = 10
b = 3.4
c = 2 + 3j
d = 0
# Write your code here.
Exercise 4.12 ([*] Strings)
For this exercise, you can use the following documentation.
A sequence is a data structure that contains items arranged in order, and you can access each item using an integer index that represents its position in the sequence. Note that Python starts counting at zero!
A string is a sequence of characters. It is useful for representing text. We make a string by using single or double quotation marks.
In this exercise, we will explore a strand of genetic material. First, check the data type of the following strand:
strand = "ACGUGCAGCUCGGGCGAUCGCGCUAAAGCGCAUUCGCGUACGAUGGGCGAA"
# Your code here
As genetic material is often represented by letters, we use strings to encode them. There are a few operations you can perform on strings that make them easy to inspect and manipulate.
As you can see from the strand above, it must be an RNA strand due to the presence of uracil. However, you may have a very long string of characters and not be able to see immediately whether there are U-bases in the strand. Write a piece of code that uses a Boolean variable to check whether there are U bases in a strand.
contains_U_bases = .. in ..
# Here, "in" is used to make Python search for a specific character in a string. If found, it returns "True".
print(type(contains_U_bases))
print(f"This strand contains U-bases and is therefore an RNA strand: {contains_U_bases}")
Now, turn this RNA strand into an mRNA strand by adding a G-cap and a poly-A-tail. Note that you can add strings together using the + operator.
G_cap =
poly_A_tail =
mRNA_strand =
Exercise 4.13 ([*] Print statement)
You can use the following documentation for this exercise.
You have already encountered some examples of the print() function. The print function allows you to read the outputs of your code. It is a versatile tool that is very useful for debugging and figuring out whether your code actually does what you think it does. In this exercise, we will explore some useful properties of the print function.
The print function itself is fairly simple. Use it to print the statement “The answer is 42”.
# Your code here
You can also use print() to print the value of a variable. Define and print a variable in the following cell.
# Your code here
You can do some simple formatting in a print statement. For example, use \n for an enter and \tfor a tab. Print this haiku in the proper form (5/7/5 syllables per line): “coding is the best / don’t let it make you feel dumb / keep calm, debug on”.
# Your code here
Finally, you can enter multiple variables and/or objects in a single print statement using commas to separate the string and the variables. In the following code, print a cohesive sentence containing all information on the following protein:
protein_name = "Sonic Hedgehog protein"
number_of_amino_acids = 462
location_encoding_gene = "Chromosome 7"
# Your code here
Formatted strings (f-strings)
To combine multiple types of variables in a string or in a print statement, we can also use f-strings. These usually make your code a bit more readable. To create an f-string, we preface the string with an f (f"..") and we put the variables between curly brackets ({variable}). For example:
print(f"The {protein_name} has {number_of_amino_acids} amino acids and can be found on {location_encoding_gene}.")
Exercise 4.14 ([*] Input)
For this exercise, you can use the following documentation.
The input() function allows you pass information to Python. Between the brackets, you can specify the prompt that Python shows you when you have to enter something.
Run the following code and provide the input that is asked for. Check the type of the input you provided.
a = input("Input a string. ")
b = input("Input an integer. ")
c = input("Input a float. ")
# write code to check the types here
As you can see above, Python always interprets your response as a string. To use the variable further, you need to convert it to the type you want. In the code below, use input() and type converting functions to get the variable type that you want. Verify that it worked using print statements.
b =
c =
You can also directly input multiple variables. In this example, we will input one integer and one float separated by space and save it as two_numbers string. To then separate the two inputs, we use a comma before the equal sign (b,c =) and the built-in function .split() which cuts up the string (as default, based on spaces). It can be used as follows: string_name.split(), so in our case b,c = two_numbers.split().
two_numbers = input("Input an integer and a float separated by a space")
b,c = two_numbers.split()
print(f"The first variable is {b} and the second variable is {c}.")
Now write the code to do the same using two lines: one line to ask for input and split the string, and the other a print line.
# your code here
Exercise 4.15 ([*] Sequences)
For this exercise, you can use the following documentation.
Tuples
A sequence is a data structure that contains items arranged in order, and you can access each item using an integer index that represents its position in the sequence. You have already encountered strings as an example of a sequence.
A tuple can be used to store multiple items in a single variable. Tuples are immutable, meaning that they cannot be changed after they have been defined. They are indicated by round brackets: (item1, item2, ..).
In this exercise, we consider a DNA sequence, which is stored as strings. Determine the GC-content of the sequence; this is the fraction of the DNA sequence made up by G or C bases. The GC-content is important to determine the stability of the DNA-molecule. We use the method .count() to find how many occurrences there are of a specific base in a string.
DNA_sequence = "GTACGAGCATGCTGG"
G_number = DNA_sequence.count("G")
# Extend the code for other bases
Knowing the number of G and C bases, we can determine the melting temperature of the DNA, i.e., the temperature at which the strands separate from one another, using the Wallace rule:
\( T_m \text{ (in }^{\circ}C\text{)} = 4 * \text{(number of Gs + number of Cs)}+ 2 * \text{(number of As + number of Ts)}\)
Calculate the melting temperature of the DNA sequence.
melting_temperature = # add your code here
print(f"The melting temperature is {melting_temperature} degrees Celsius")
Store the variables with information on the DNA (melting temperature, GC content) in a tuple to have all of it in one place:
sequence_information = (.., ..)
Run the following code to check whether your sequence_information is correct:
from check_08 import *
check_08(DNA_sequence, sequence_information)
Imagine we want to raise the melting temperature and increase the stability. To do so, we would have to increase the GC-content. Add 4 G or C bases to the original string and calculate the new melting temperature.
new_DNA_sequence =
new_melting_temperature =
Index the tuple sequence_information to retrieve the melting_temperature of the initial strand. By how much did the melting temperature increase?
# Your code here
Exercise 4.16 ([*] Lists)
For this exercise, you can use the following documentation.
A sequence is a data structure that contains items arranged in order, and you can access each item using an integer index that represents its position in the sequence. You have already encountered strings as an example of a sequence.
Lists allow you to store multiple items in a single variable. They have a defined order, which allows you to retrieve items based on their position in the list. Contrary to tuples, however, lists are mutable, which means you can easy change, add, or remove items.
In this exercise, we’ll explore the usefulness of lists by looking at a list containing all 20 naturally occurring amino acids:
amino_acids = [
"Methionine", "Histidine", "Threonine", "Arginine", "Alanine",
"Glycine", "Tyrosine", "Proline", "Asparagine", "Valine",
"Phenylalanine", "Leucine", "Glutamine", "Cysteine", "Aspartic acid",
"Isoleucine", "Serine", "Tryptophan", "Lysine", "Glutamic acid"
]
First, sort this list alphabetically using built-in function sorted().
amino_acids = # Add your code here
Now, determine the index of Glutamine (hint: see the documentation) and use that to retrieve it from the list:
index_glutamine =
glutamine = amino_acids[..]
print(glutamine)
Define a list of all amino acids starting with an A by using slicing, i.e., indexing of multiple items. You can slice multiple items as follows: list_name[start:stop] - the item with index start will be included, whereas the item with index stop will not be included.
amino_acids_starting_with_an_A =
print(amino_acids_starting_with_an_A)
Add the amino acid “Selenocysteine”, the Se-analogue of cysteine, to the list. Make sure that your final list is again sorted alphabetically!
# Add your code here
You can check whether a specific item is in a list using a Boolean variable. Use input() and the Boolean variable is_this_an_amino_acid to check whether your input string is in the extended list of amino acids.
your_amino_acid = input() # make sure that this variable is of the correct type
is_this_an_amino_acid = .. in ..
print(f"{your_amino_acid} is an amino acid: {is_this_an_amino_acid}")
Exercise 4.17 ([*] Dictionaries)
For this exercise, you can use the following documentation.
A dictionary is associative: it links a category to its components, or in Python-language, a key to its values. You can change a dictionary after it has been defined, i.e., it is mutable or changeable.
Make a dictionary which contains the words of a sentence as keys and the next word as value. For example, the sentence “Luke, I am your father” would correspond to the following dictionary:
example_sentence_dictionary = {"Luke" : "I",
"I" : "am",
"am" : "your",
"your" : "father"}
Make a similar dictionary for the sentence: “if you can do it”. Do so by defining an empty dictionary and progressively adding items.
sentence_dictionary = {} # Define an empty dictionary
# You can add an item to the dictionary in the following way:
sentence_dictionary["if"] = "you"
Now if we slightly change our sentence, we want to be able to remove a key and its value. The new sentence is: “you can do it”.
Change the existing dictionary to reflect the change in the sentence. Use the .pop() method to remove an item. Between the brackets, you only have the include the key; .pop() will then remove both the key and its value.
# Add your code here
print(sentence_dictionary)
Now imagine you want to see which word comes after “do” in the sentence. You can retrieve a value corresponding to a specific key by “indexing” the dictionary with that key. Try this yourself to see which word comes after “can”. Also check what happens when you enter a key that is not in the dictionary at all; what kind of error do you get?
word_after = sentence_dictionary["do"]
print(word_after)
Making a sentence dictionary becomes more difficult if you have words that appear in the sentence multiple times. As a value can only contain one variable, you will need to use a list to have multiple words associated to one key. For example, the sentence “It’s the eye of the tiger” would become
sentence_dictionary = {"It's" : ["the"],
"the" : ["eye", "tiger"],
"eye" : ["of"],
"of" : ["the"]}
Make such a dictionary yourself of the sentence “I drink coffee and I eat a pastry”. Again, start from an empty dictionary and progressively add items. Remember: if you want to add an item to a list, you can use the method .append().
sentence_dictionary = {}
sentence_dictionary["I"] = ["drink"]
Exercise 4.18 ([**] Sets)
For this exercise, you can use the following documentation.
A set is a variable that, like a tuple, is immutable. Furthermore, it is not ordered (so you cannot retrieve items based on their index), and there cannot be any duplicates in a set.
In this exercise, we will explore some of the applications of sets. First consider the following list with all individual types of cells found in a (very small) patient sample of a tumour.
tumor_cells = ['T cell', 'Dendritic cell', 'T cell', 'Dendritic cell', 'Cancer cell', 'B cell',
'Macrophage', 'Cancer cell', 'Cancer cell', 'Macrophage', 'B cell', 'Fibroblast', 'T cell',
'T cell', 'Cancer cell', 'Macrophage', 'Dendritic cell', 'Tumor-infiltrating lymphocyte',
'Tumor-infiltrating lymphocyte', 'Macrophage', 'Cancer cell', 'Endothelial cell',
'Cancer cell', 'Fibroblast', 'Dendritic cell', 'Endothelial cell',
'B cell', 'Cancer cell', 'Tumor-infiltrating lymphocyte', 'Fibroblast', 'T cell',
'Endothelial cell', 'T cell', 'Macrophage', 'Endothelial cell', 'Macrophage', 'B cell', 'Cancer cell',
'Cancer cell', 'T cell', 'Cancer cell']
First check how many items there are in this list by using the built-in function len().
number_of_cells = # Put your code here
Then, check how many cancer cells there are in the sample using count(), and what percentage of all cells is made up by cancer cells.
# Your code here
Now, to get a better overview of the cells that are actually present, we would like to remove all duplicates. The easiest way to do this is by turning the list into a set using set(). Print the result.
unique_cell_types = set(tumor_cells)
From this list of cell types, we see three categories: immune cells (T cell, B cell, Macrophage, Tumor-infiltrating lymphocyte, Dendritic cell), stromal cells (Endothelial cell, Fibroblast) and cancer cells. Create a dictionary with the category as key and a list of the corresponding cell types as value.
categories_of_cells =
You receive a second sample of cells. You want to figure out whether there are any types that are in the first sample (from now on referred to as sample A) but not in the second sample (sample B). Fortunately, you can apply set theory in Python on sets, meaning that you can find their difference (using -), but also their intersection (using &), their union (using |), their difference (using ^) and more.
Find the cell types that differ between sample A and sample B.
tumor_cells_sample_B = ['Neutrophil', 'Dendritic cell', 'T cell', 'Dendritic cell', 'Cancer cell', 'Neutrophil', 'Pericytes', 'Cancer cell',
'Cancer cell', 'Pericytes', 'Neutrophil', 'Fibroblast', 'T cell', 'T cell', 'Cancer cell', 'Pericytes', 'Dendritic cell',
'Tumor-infiltrating lymphocyte', 'Tumor-infiltrating lymphocyte', 'Pericytes', 'Neutrophil', 'Endothelial cell',
'Cancer cell', 'Fibroblast', 'Dendritic cell', 'Neutrophil', 'B cell', 'Cancer cell', 'Tumor-infiltrating lymphocyte',
'Fibroblast', 'Neutrophil', 'Endothelial cell', 'T cell', 'Pericytes', 'Endothelial cell', 'Pericytes', 'B cell',
'Cancer cell', 'Cancer cell', 'T cell', 'Neutrophil']
# Your code here
Finally, going back to our set of unique cell types in sample A, we note that we’re not terribly interested in the stromal cells (i.e., the fibroblasts and the endothelial cells). Remove these from the cell types.
# Your code here
Exercise 4.19 ([**] Organelles)
You receive the radius of a typical human skin cell, as well as two dictionaries, one with the radii of various organelles and one with the number of these organelles typically found in a skin cell. Write a piece of code that calculates the volume percentage each of these organelles takes up in the cell. You may assume that the organelles are spherical.
Store the volume percentages in a dictionary named volume_percentages with the same keys as the original two dictionaries. Do not round the percentages.
pi = 3.1415
cell_radius = 40 # micrometers
organelle_radius = {"nucleus" : 15,
"mitochondrion" : 3,
"golgi" : 5e-1,
"ribosome" : 1e-3} # all in micrometers
organelle_number = {"nucleus" : 1,
"mitochondrion" : 200,
"golgi" : 1,
"ribosome" : 10e7}
# Start your solution here
Solution to this exercise
{‘nucleus’: 5.2734375, ‘mitochondrion’: 8.437500000000002, ‘golgi’: 0.0001953125, ‘ribosome’: 0.00015625}
Exercise 4.20 ([**] Mutating a genome)
For this exercise, take a look at the following information.
An organism with a very small genome has 3 chromosome pairs of 6, 8, and 10 bases. You get a list of these chromosomes in order (i.e., the first two entries are your chromosome 1 set, the second two entries are the chromosome 2 set, etc.). You want to mutate chromosome set 3 such that the 7th base becomes a “G” in both copies. Use square brackets to access chromosomes and nucleotide bases.
genome = ["CGTACG", "GTCTAT",
"ATTCTACA", "TGCAAGCT",
"TGCGCACTAC", "ATGCTATCTG"]
# Start your solution here
Exercise 4.21 ([*] Watermelons (error types I))
The code below will give an error. Figure out what the type and the source of the error is and fix it.
price_watermelon_per_kilo = 1.97 # euros
number_of_small_watermelons = 20
number_of_big_watermelons = 15
weight_small_watermelon = 2.7 # kilograms
weight_big_watermelon = 4 # kilograms
total = price_watermelon_per_kilo * (number_of_small_watermelons * weight_small_watermelon +
number_of_big_watermelons * weight_big_watermelon)
total = round(total, 2)
print("The total price is €" + total)
Exercise 4.22 ([*] RNA to mRNA (error types II))
This code checks the length of an RNA strand and the maximum potential length of its corresponding protein. Next, it turns the RNA strand into an mRNA strand and checks whether this was successful.
The code below will give an error. Figure out the type and the source of the error and fix it.
RNA_strand = "ACGACCGGGAAGCACGGACCGGAAGGCGCGACCAAGGCGCACGGGACGUGAACGCCGACGGGACCGGCGACGAC"
# Determination of characteristics
length_of_RNA_strand = len(RNA_strand)
mod_RNA_bases = lenght_of_RNA_strand % 3 # Here, we calculate how many bases are not part of a triplet
maximum_potential_length_protein = (length_of_RNA_strand - mod_RNA_bases) / 3
print(f"The maximum potential length of the protein is {maximum_potential_length_protein:.0f} amino acids.")
# Transformation into mRNA
G_cap = "G"
poly_A_tail = "AAAAAAAAAAAAAAAA"
mRNA_strand = G_cap + RNA_strand + poly_A_tail
successfully_G_capped = (mRNA_strand[0] == "G") # Looks at the first character in the string
successfully_A_tailed = (mRNA_strand[-5:-1] == "AAAA") # Looks at the final four characters
print(f"The RNA strand has been successfully transformed into mRNA: {successfully_A_tailed and successfully_G_capped}")
# The word "and" only returns True if both conditions are True.
Exercise 4.23 ([**] Measurements (error types III))
We want to take a list of ten measurements and calculate the average and range.
The code contains two mistakes. One of these will give an error, the other is a conceptual mistake. Figure out the mistakes and fix them; furthermore, figure out what type of error this code gives.
measurements = [1.5, 3.0, 2.7, 1.1, 7.8, 5.4, 3.7, 4.4, 9.6, 5.6]
measurements = sorted(measurements)
print(f"The sorted list of measurements is the following: {measurements}")
number_of_measurements = len(measurements)
highest_value = measurements[number_of_measurements]
lowest_value = measurements[1]
range = highest_value - lowest_value
average = sum(measurements) / number_of_measurements
print(f"The range of the measurements is {range:.2} whereas the average of the measurements is {average:.2}.")
Bonus question: What does the {range:.2} do in the print statement above?
Exercise 4.24 ([**] Air bubble velocity)
Air bubbles “trapped” in a viscous liquid will slowly rise to the surface. The velocity of these bubbles can be calculated by solving the force balance equation:
where \(k\) is a drag coefficient, \(v\) the velocity of the bubble, \(\rho_V\) the density of the air bubble, \(\rho_w\) the density of the liquid, \(V\) the volume of the bubble (assumed to be spherical), and \(g\) the acceleration due to gravity. All numbers should be given using standard SI units (kg, m, s, etc.).
The outline code to calculate the air bubble velocity is given below.
Your task is to:
Complete the code, where the diameter is asked as input value by the user in cm.
Calculate the volume of the spherical air bubble. Use 3.14 as the value of \(\pi\).
Print the velocity of the air bubble and the associated unit, e.g., “The velocity is: … m/s.”.
It is considered good practice that the units are given directly after the numbers (as shown with \(g\)). Provide the three missing units in the code below.
k = 0.25 # Add unit
rho_V = 1.29 # Add unit
rho_w = 0.998 # Add unit
g = 9.81 # m/s^2
# Your code for defining d and V
# Your code for calculating the air bubble velocity and printing output
Exercise 4.25 ([**] Bacterial growth (error types IV))
Imagine you are in the lab growing bacterial colonies. As bacteria are allowed to divide, they will accumulate DNA damage. This code calculates the average number of mutations that have occurred in a bacterium over a specific amount of time.
The code contains two mistakes. Both these mistakes will give an error. Figure out what these errors mean and fix the mistakes in the code.
mutation_rate = 1e-10 # This means that for every 10 billion bases copied, there will be one mutation
genome_size = 4e6 # This is approximate, since it - technically speaking - depends on the species
initial_number_of_bacteria = 1e3
# The generation time is the time a specific species needs to divide, i.e. double its population.
generation_time_various_bacteria = {"C. perfringens" : 10
"E. coli" : 20
"S. enterica" : 30
"P. aeruginosa" : 30
"V. cholerae" : 40}
bacterial_species = input("What bacterial species are you working with? ")
time_elapsed = int(input("For how many minutes have the bacteria been allowed to divide?"))
# The following two lines of code checks whether the growth of bacteria has plateaued by setting an upper limit to the division time.
time_elapsed = max(time_elapsed, 1000)
generation_time = generation_time_various_bacteria["bacterial_species"] # We take the generation time of the entered species
number_of_divisions = round(time_elapsed/generation_time) # We calculate how many divisions have taken place in the specified time
total_number_of_bacteria = initial_number_of_bacteria * 2**number_of_divisions # We find the total number of bacteria after these divisions
number_of_mutations_per_bacterium = number_of_divisions * genome_size * mutation_rate # We find the number of mutations so far
print(f"On average, the number of mutations per bacterium is {number_of_mutations_per_bacterium:.2}, "
f"there will be one mutation in every {(1/number_of_mutations_per_bacterium):.2f} bacteria.")
Exercise 4.26 ([***] Atomic force microscopy (AFM) data)
You are studying a protein in different conformations under the AFM. Your raw data is a list of numbers; every number corresponds to a specific conformation. You also receive an image (see below) containing the names of the conformations that the numbers correspond to. Convert the list of numbers to a dictionary which has the names of the conformations as key and the number of times that this conformation has been found as value.
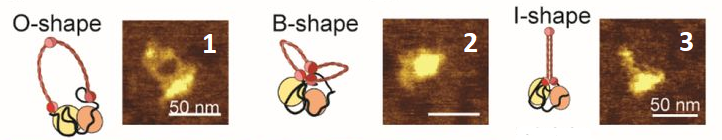
conformations = [2, 1, 3, 2, 1, 3, 2, 2, 1, 1, 3, 3, 1, 2, 2, 1, 3, 1, 1, 2, 3, 3, 2, 1, 2, 2, 3, 1, 1, 3, 3, 2, 1, 2, 1, 1, 3, 3, 2, 2, 1, 3, 1, 2, 2, 3, 3, 1, 2, 1, 1, 3, 3, 2, 1, 1, 2, 3, 3, 1, 2, 2, 3, 1, 1, 2, 2, 3, 3, 1, 1, 2, 3, 3, 1, 1, 2, 2, 1, 3, 3, 1, 2, 2, 3, 1, 1, 3, 2, 2, 1, 1, 3, 2, 2, 1, 1, 3, 2, 1, 3, 1, 1, 2, 3, 2, 1, 1, 3, 3, 2, 1, 2, 1, 3, 3, 2, 1, 1, 3, 2, 2, 1, 3, 3, 1, 2, 1, 1, 3, 2, 1, 3, 1, 2, 2, 3, 1, 1, 3, 2, 2, 1, 1, 3, 3, 1, 2, 1, 3, 2]
# Your code here
Exercise 4.27 ([***] Different clock)
We have 24 hours per day, 60 minutes per day, and 60 seconds per minute. A new clock is proposed: 10 tiks per day, 100 taks per tik, and 100 toks per tak. Write a function to convert our regular 24:60:60 time to tik-tak-tok’s 10:100:100 time.
First, calculate how many seconds there are in a day:
# Your code here
Now calculate how many seconds one ‘tak’ is:
# Your code here
Use this information to reprogram a digital clock from the max 23:59:59 in hours:min:seconds, to max 09:99:99 in tik:tak:tok.
hms = [15, 26, 39]
# put your code here
tik_tak_tok =
Run the following code to check whether your solution is correct:
from check_19 import *
check_19(hms, tik_tak_tok)
Exercise 4.28 ([***] Inverting a codon dictionary)
Below, you’ll find a dictionary showing which codons (only the ones starting with an A for simplicity purposes) correspond to which amino acid. In this dictionary, the amino acid is the key of each entry, whereas the corresponding codons are the values.
In the exercise, you must invert this dictionary, such that the keys are the values and the values are amino acids. Note that you need loops for this exercise, which will only be covered later in the book, so we recommend only trying this exercise if you have some previous programming experience.
amino_acid_to_codon_A = {
'Lysine': ['AAA', 'AAG'],
'Asparagine': ['AAC', 'AAU'],
'Threonine': ['ACA', 'ACC', 'ACG', 'ACU'],
'Arginine': ['AGA', 'AGG'],
'Serine': ['AGC', 'AGU'],
'Isoleucine': ['AUA', 'AUC', 'AUU'],
'Methionine': ['AUG']
}
codon_to_amino_acid_A = {}
for amino_acid, codons in amino_acid_to_codon_A.items():
# Your code here
Now that you have inverted it, you can use the new dictionary to translate a sequence of codons! Try it out with the following sequence:
sequence = "AUGAUAAUUAAGAAGAAAACCAGGAGCAUAAUUAAAACAACG"
# Your code here