9.2. Files with NumPy#
Interactive page
This is an interactive book page. Press launch button at the top right side.
Let’s explore NumPy functions for working with data files.
9.2.1. Loading, opening, and reading a file#
To load data from a text file into Python using NumPy, you can use its function np.loadtext(). To learn more about this function, use ?np.loadtxt or see documentation. Let’s try it out.
File location and common bugs
The code below works when a file (such as v_vs_time.dat) is located in the same folder as your Python script. If your file is somewhere else, e.g., one folder up, you have to use your knowledge of navigating directories in Terminal. You would then use data = np.loadtxt("../v_vs_time.dat") where ../ indicates the location of your file.
In general, if your file won’t load into Python, often you’re either looking for it in the wrong directory, or have a typo in either path or file name. For this, the terminal in VS Code can come handy with pwd and ls commands.
import numpy as np
data = np.loadtxt("v_vs_time.dat")
print(data)
Here we loaded the data from the v_vs_time.dat file and printed it. While not obvious from the data itself, the file name suggests what this data is about: measurements of voltage as a function of time. In this case, the data is recorded in two columns of numbers in a text file, where numbers in two columns are separated by a space. The first number is the time of the measurement (in seconds) and the second is the measured voltage (in volts).
This is an example of a DSV (delimiter separated value) file - the delimiter (i.e., the thing that separates the values in two columns) is a space . Another common delimiter is a comma , and such files are CSV (comma separated value) files. CSV is a common “export” format from spreadsheets like Excel. A third common delimiter is Tab with TSV (tab separated value) files (also available as an export option from some spreadsheets). When values are separated by Tab characters, this shows up in Python strings as a special character \t.
New line character
Similar to \t for Tab, Python has a new line character \n. For instance, if you use this print statement:
print("Hello world! \nGoodbye!")
it prints in two lines:
Hello world!
Goodbye!
The NumPy function np.loadtext() can handle any type of delimiter: the default is any whitespace (Tab or space), but this can also be changed using the delimiter= keyword argument of np.loadtext(). np.loadtext() also does not care about the file extension, e.g., it is fine if a CSV file does not have a .csv extension (it doesn’t even have to have an extension). Files containing ASCII text data often carry the extension .dat.
What if the file above was a comma separated file, and it also had a header describing what the columns actually are? How would we import it?
A glance into a file
When working with a new text file, how do you check what’s the delimiter, and what the file looks like in general? To get a quick glance at the file’s content: open your terminal, navigate to the directory with the file, and then read the file using less file_name. This can help you determine how to use np.loadtext() to successfully load your file.
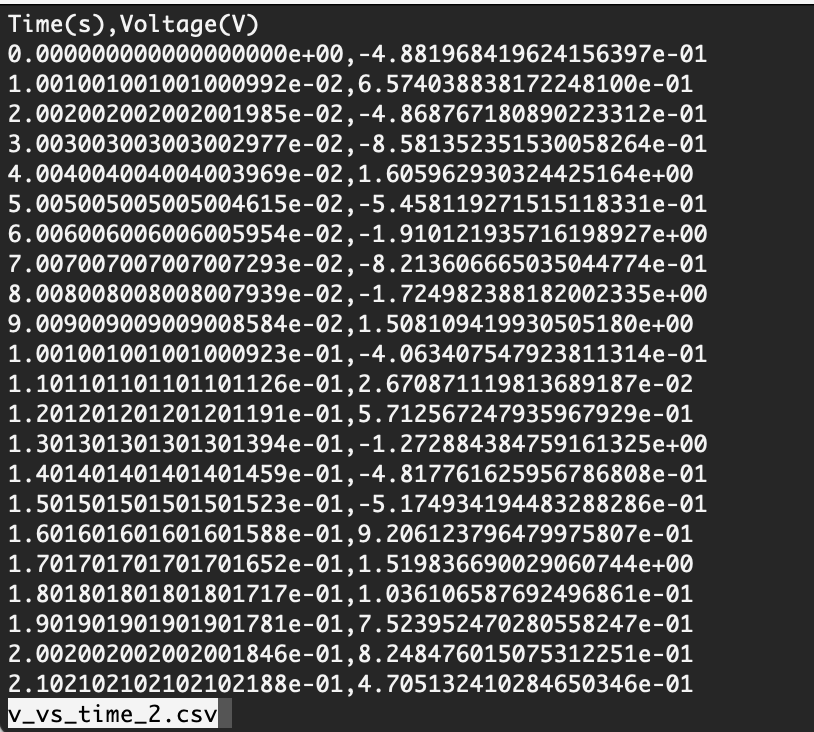
Fig. 9.2 v_vs_time file in CSV format and a header, as viewed in terminal with the less command.#
import numpy as np
# Set delimiter to comma ";" and skip first (header) row
data_2 = np.loadtxt("v_vs_time_2.csv", delimiter=",", skiprows=1)
print(data_2)
Here we saw how to “tweak” np.loadtxt() so that it works perfectly for our second file! As a general note: it’s very useful to explore which arguments a Python function can take.
In both cases above, we assigned the return value of np.loadtxt() to a variable (data and data_2), which are NumPy arrays (you can check this with the type command). When we printed our variables, we saw this:
[[ 0.00000000e+00 -4.88196842e-01]
[ 1.00100100e-02 6.57403884e-01]
[ 2.00200200e-02 -4.86876718e-01]
...
[ 9.97997998e+00 2.11430345e+01]
[ 9.98998999e+00 1.94693126e+01]
[ 1.00000000e+01 1.82114232e+01]]
The dots ... suggest that Python ommitted some lines there in the middle (this happens when the data is large, otherwise it could completely clutter your screen). So how large is our array data? We can find out with np.shape():
print(np.shape(data))
Therefore, when np.loadtxt() loads a file, it returns a 2D NumPy array with shape (n,m), where n is the number lines in the file and m is the number of columns (here, we have 1000 rows and 2 columns).
As mentioned above, the first column represents the time in seconds when the measurement was taken, and the second column represents the measured voltage in volts. We will typically want to extract these into two vectors t and v. We can do this using slicing:
t = data[:,0]
v = data[:,1]
We can choose to look at the first ten points, and see that we have successfully loaded the data from the file and extracted it into vectors.
print('t = ', t[0:10])
print('v = ', v[0:10])
You should always double-check that the data was loaded correctly by comparing it with the file opened in terminal, Excel, or another application. It’s also possible to open the file in Python and print its contents line by line - let’s see how to do this for the first ten lines:
with open("v_vs_time.dat") as file:
for i in range(10):
print(file.readline())
9.2.2. Saving a file#
We can also save data using NumPy’s np.savetxt(). To do this, we first have to make sure that the data is a NumPy array of the correct shape.
Let’s take a look at an example where we calculate the square of the measured voltage (the vector from above) and save this back into a new file:
v_squared = v**2
To “pack” the new vector v_squared and the unchanged time vector t together into a matrix like the one returned by np.loadtxt(), we can first create a 2D matrix of the correct size and then use slicing with an assignment operator = to give the columns the correct values:
# Create an empty array
matrix_to_save = np.zeros([len(v), 2])
# Fill columns with data
matrix_to_save[:,0] = t
matrix_to_save[:,1] = v_squared
Now we are ready to use np.savetxt(), which (unless specified otherwise) saves the file in the current directory:
np.savetxt("vsquare_vs_time.dat", matrix_to_save)
This creates a file in your workspace, which you can then open to see what’s inside it.
import micropip
await micropip.install("jupyterquiz")
from jupyterquiz import display_quiz
import json
with open("questions6.json", "r") as file:
questions=json.load(file)
display_quiz(questions, border_radius=0)
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[10], line 1
----> 1 import micropip
2 await micropip.install("jupyterquiz")
3 from jupyterquiz import display_quiz
ModuleNotFoundError: No module named 'micropip'