6.5. Exercises#
Each exercise carries the difficulty level indication:
[*] Easy
[**] Moderate
[***] Advanced
The highest difficulty level is meant for students who wish to challenge themselves and/or have previous experience in programming.
Exercise 6.12 ([*] Boolean logic)
Note: this exercise contains an introduction, which is absent from the downloadable exercise (.py) file.
Introduction
Boolean logic
Boolean logic is a fundamental underlying technique for computers. It works based on two values - true and false.
In the computer, this is represented as 1 and 0, respectively. In Python, these are the bool types (True and False).
There are a number of basic operations (Boolean operators):
AND
OR
NOT
We’ll go through each one individually first.
The basic operators:
AND returns true (1) if and only if both of the given values are true.
The AND operator is represented in Python by the keyword and.
OR returns true if either of the inputs are true. This also means that if both inputs are true,
it will also return true. The OR operator is represented in Python by the keyword or.
NOT returns the opposite of the given value.
For instance, if you have the value a = True, not a will return False.
The NOT operator is given in Python by not.
Combinations of operators
The basic operators can be combined to produce more advanced behaviors. For instance, if you need three values to all be true for something to happen, then it may be combined in a statement something like (a and b) and c.
It may look clunky, but it does work.
In a more real-world example, say you are modelling a cell dividing. To divide, it needs to check that a number of things are correct:
Has the DNA been copied successfully?
Is there any cell damage?
Is there enough space to divide? This can be checked by combining everything into one large
andstatement.
Truth tables
Truth tables are a tool that presents in an organized way the outcome of a statement depending on the inputs. It is a useful way for organizing or figuring out the result of many inputs into a complex statement
In this set of exercises, we will combine basic Boolean operators to define when certain biological events are allowed to take place.
Exercise A: Cell cycle G1 checkpoint
Define three variables - DNA_damaged, E2F_present, and not_touching - and assign
Boolean values to them. The statement can_pass_g1 should be true if there is
enough space, no DNA damage, and no E2F proteins. You can try to assign
different values to the three variables and see how these affect the outcome
(can_pass_g1).
Use this as a guideline:
DNA_damaged- true if damagedE2F_present- true if presentnot_touching- true if it is not overcrowded
# Your code here
can_pass_g1 = # Your code here
Exercise B: Cell cycle G2-M checkpoint
Define two variables - DNA_damaged and cyclinB_present - and assign Boolean values
to them. The statement can_pass_g2m should read true if there is no DNA damage
and if cyclinB is present. You can try to assign different values to the two
variables and see how these affect the outcome (is the cell allowed to pass the
cell cycle checkpoint).
Use this as a guideline:
DNA_damaged- true if damagedcyclinB_present- true if present
# Your code here
can_pass_g2m = # Your code here
Exercise C: Enzyme production
Define two Boolean variables - MetA_present and MetB_present - and assign value to
them. Based on these variables, define make_enzyme. The cell should start
to produce the enzyme if metabolite A or metabolite B is present, but not both.
How does changing the values of MetA_present or MetB_present affect the outcome?
Use this as a guideline:
MetA_present- true if presentMetB_present- true if present
# Your code here
make_enzyme = # Your code here
Exercise D: (Optional) Cancer
Rewrite Exercise A so that the behaviour would imitate that of cancer.
# Your code here
Exercise 6.13 ([*] Logical statements: debugging)
Note: this exercise contains an introduction, which is absent from the downloadable exercise (.py) file.
Introduction
Logical statements
In the previous exercise, we covered Boolean logic - combining true and false statements to generate logic. The next logical question is then: “how to generate these true-false statements?” This is done using logical statements.
Numerical/value comparison
This type of logical statements is the simplest one, where we essentially compare a value (the variable containing that value) to another one. There are the following operators:
==- Equals (note the double equal)>- Greater than<- Less than!=- Is not equal to.!often acts as a NOT operator in programming>=- Greater or equal to<=- Less than or equal to
With Python, it is possible to add a lot of things to the comparison. For instance, doing simply:
str_a = "this is a test"
str_b = "this is a test"
is_the_same = (str_a == str_b)
will result in is_the_same being True.
You will use these types of comparisons in most of your coding.
Other ways to generate True/False
Boolean True/False may also be given by some other things:
Non-Boolean variables can by themselves be interpreted as Booleans, e.g., numerics (
float,int) if they are equal to 0 are interepreted asFalse.inchecks if an item is contained within a set-like Python type, e.g.,x in array_ytests if the value ofxis inarray_yand returnsTrueif it is. It also works for strings.A command or function can itself return
TrueorFalse, which may be useful for catching errors or exceptions.
Exercise A: Predict True or False
For the three pieces of code below, predict whether they will return True or False
without running the code. To check whether your prediction is correct, run the
code with the needed print() statements.
# Code 1
gfp_prot_seq = ""
pred1 = bool(gfp_prot_seq)
# Your prediction for pred1 = True or False?
# Code 2
gfp_prot_seq = "MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK"
pred2 = bool(gfp_prot_seq)
# Your prediction for pred2 = True or False?
# Code 3
animal = "rabbit"
mammals = ["giraffe", "elephant", "lion", "rabbit", "guinea pig", "vole", "shrew", "human", "cat", "dog"]
reptiles = ["crocodile", "iguana", "alligator", "boa", "cobra", "python", "anole", "turtle", "tortoise", "tuatara"]
pred3 = animal in mammals
# Your prediction for pred3 = True or False?
Exercise B: Code debugging
The statements in each of the three pieces of code below should be true. Understand why they are not, and fix the code.
# Code 1
x = 1.4 * 1.5
print(f"Is x = 2.1? {bool(x == 2.1)}")
# Code 2
y = 'a' + 7
test_ar = ['a', 'b', 'c', 'd', 'e', 'f', 'g']
print(f"Is y in the test_ar? {bool(y in test_ar)}")
# Code 3
test_ar = ['a', 'b', 'c', 'd', 'e', 'f', 'g']
z = 7
print(f"Is z longer than the test array? {bool(z > len(test_ar))}")
Exercise 6.14 ([**] Lamp: debugging)
The code below is the implementation of the following algorithm:
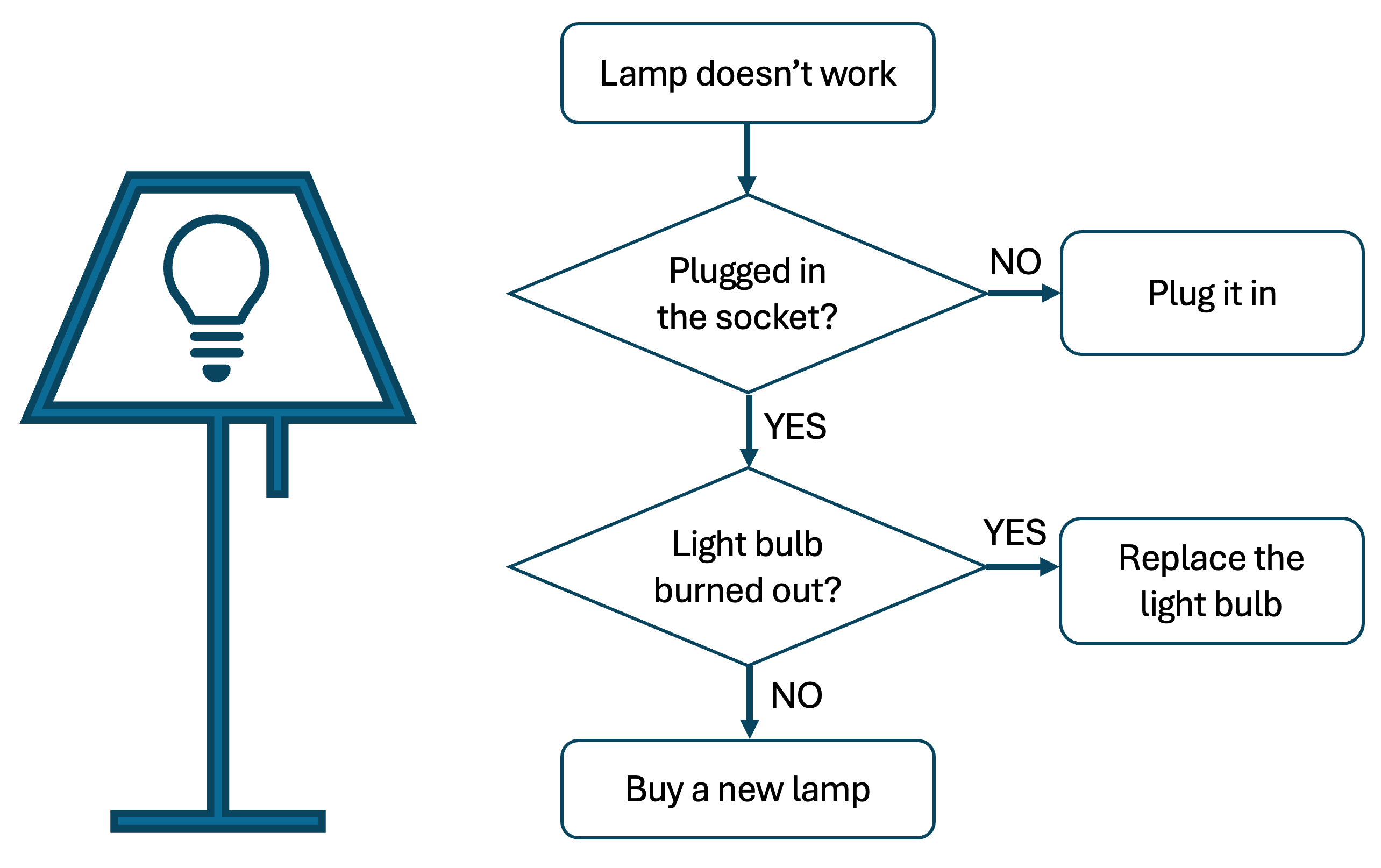
However, both logical and syntax bugs are introduced into the code. Try to fix them all and make a functional code implementation of the outlined algorithm.
Hint: there are six bugs.
lamp_working = "yes"
plugged_in = "no"
burned_bulb = "yes"
if lamp_works == no:
if plugged_in == "no":
print("Plug it in!")
else:
if burned_bulb = "yes":
print("Buy a new lamp :(")
else:
print("Replace the light bulb!)
else:
print("All is good :)")
Exercise 6.15 ([*] DNA sequence and if/else/elif)
Note: this exercise contains an introduction, which is absent from the downloadable exercise (.py) file.
Introduction
The if statement
The if statement is the most basic flow control statement; it indicates that if something is true, then the program should do something.
a = True
if a:
print("a is true")
The part immediately following if is a logical statement.
Any code that is indented will be run if the logical statement is true. If not, it will be skipped.
Some additional notes on what can be put in as a logical statements:
A single Boolean value can be put in without comparison to
TrueorFalse.Lists, strings, and tuples can also be put into an
ifstatement. They are interpreted asFalseif they are empty,Trueif they are not.Any numeric value (
int,float, etc.) equal to 0 is also interpreted asFalse.
The else statement
Sometimes you want the computer to do something else in the case the original if statement is not true.
This is where else is used.
if a is True:
print("a is true")
else:
print("a is false")
In this example, it will printed that a is false if a is not true.
Note that this code:
test_var = True
if test_var:
print("The test variable is true")
else:
print("The test variable is false")
will behave differently than this one:
test_var = True
if test_var:
print("The test variable is true")
print("The test variable is false")
In the latter case, the computer does not “know” that the second print line should be exclusive with the first print.
The elif statement
Sometimes, the computer needs to make more than one decision at a time.
This can be written:
if a is True:
# do something
else:
if b is True:
# do something else
else:
if: c is True
# do a different thing
else:
# do another thing
This uses a lot of indentation and generally, is not very pretty.
elif combines the else and if into a single keyword and keeps everything on the same indent.
It makes things much neater, and a chain of decisions can be made.
The above example, for instance, becomes:
if a is True:
# do something
elif b is True:
# do something else
elif c is True:
# do a different thing
else:
# do another thing
(Advanced reading) Match Case
In some cases, you may want to check that a single variable is one of many values.
Say, for example, you want the program to behave differently depending on the cell_type.
This can be done like this, with a lot of if/elif statements:
cell_type = ""
if cell_type is "neuron":
# do neuron things
elif cell_type is "skin":
# do skin things
elif cell_type is "muscle":
# do muscle things
elif cell_type is "stomach":
# do stomach things
match case allows you to check all of these cases in a neater way:
match cell_type:
case "neuron":
# do neuron things
case "skin":
# do skin things
case "muscle":
# do muscle things
case "stomach":
# do stomach things
case _:
# do nothing
Note the last case _: - the underscore will catch anything that does not match any earlier cases.
It is the default, and typically used for some error or unexpected input.
The match case also lets you do more advanced pattern matching, which is beyond the scope of this book.
This set of exercises is meant as practice on using if, else, and
elif in Python.
These will be done on DNA sequences, or rather strings that represent DNA sequences. This may be helpful in the future in bioinformatics.
Exercise: Pseudocode
For each of the exercises below, start by writing pseudocode on a piece of paper. Only once you’re done with pseudocode should you start working on Python code in VS Code.
Exercise A: DNA subsequence
Write an if statement to check if a DNA subsequence is in a longer sequence.
Store the outcome in the Boolean variable in_seq and print the value of in_seq.
You do not need to use any loops.
You can define DNA_subsequence on your own. Check that your code works both
for subsequences that are and are not contained in the sequence.
DNA = "GGGACATATGCTCCGATTAAAGCGCATGATAGCCCGATAGGTAGATAGGGGTTCACGACG"
DNA_subsequence = # Your code here
# Your code here
Exercise B: DNA subsequence II
Repeat the above, but also use an else statement to set in_seq to false if the
DNA subsequence is not contained in the sequence.
You do not need to use any loops.
You can define DNA_subsequence on your own. Check that your code works both
for subsequences that are and are not contained in the sequence.
DNA = "GGGACATATGCTCCGATTAAAGCGCATGATAGCCCGATAGGTAGATAGGGGTTCACGACG"
DNA_subsequence = # Your code here
# Your code here
Exercise C: Handling multiple sequences
What if we want to check for presence of two different subsequences in our DNA
sequence? Write code that informs the user through print statements whether
subsequence_1, subsequence_2, none, or both of them are present in the DNA
sequence. You do not need to use any loops.
DNA = "GGGACATATGCTCCGATTAAAGCGCATGATAGCCCGATAGGTAGATAGGGGTTCACGACG"
DNA_subsequence_1 = "ATGCTCCG"
DNA_subsequence_2 = "TAGGTAGATATGGGTTC"
# Your code here
Exercise 6.16 ([**] DNA and RNA)
According to the central dogma in molecular biology, genetic information in biological systems flows from DNA to RNA to protein. DNA is a double-stranded molecule containing two complementary strands. To make mRNA, a part of a DNA strand is transcribed, and the resulting mRNA has nucleotides complementary to the DNA template (note: RNA contains uracil instead of thymine). To produce a protein, ribosomes then use codons (sets of three nucleotides) on mRNA, which are matched with their complementary anticodones on aminoacid-carrying tRNAs.
Exercise: Pseudocode
For each of the exercises below, start by writing pseudocode on a piece of paper. Only once you’re done with pseudocode should you start working on Python code in VS Code.
Exercise A: DNA strands
Find the strand complementary to the given DNA strand. Note that the input DNA string may not be only made of A, T, G, and C. The program should be able to handle this: represent a failed match as X.
Store the output string in DNA_complement variable.
DNA = "GPAGAGATTTYGTGACAGCATABGACGAGATTTTAGATACCCGATATATAGAHCG"
# Your code here
Exercise B: Transcription
Transcribe the given DNA sequence into an mRNA sequence. Note that the input DNA string may not be only made of A, T, G, and C. The program should be able to handle this: represent a failed match as X.
Store the output string in mRNA variable.
DNA = "GPAGAGATTTYGTGACAGCATABGACGAGATTTTAGATACCCGATATATAGAHCG"
# Your code here
Exercise C (Bonus): Match case
Do the same as in Exercise B, but use match case.
DNA = "GPAGAGATTTYGTGACAGCATABGACGAGATTTTAGATACCCGATATATAGAHCG"
# Your code here
Exercise 6.17 ([**] Green fluorescent protein (GFP))
Note: this exercise contains an introduction, which is absent from the downloadable exercise (.py) file.
Introduction
for loop
Very often one needs to do something multiple times, or to multiple objects.
This is where loops become useful.
There are two types within Python - for and while.
We will work with for loops first.
for loops work on a fixed number of iterations: they take some variable and run it over the same block of code multiple times.
Very often the variable is also changed in between runs (iterated over).
Iteration with range()
A standard for loop may look like the following:
for x in range(n):
print(x)
range() defines the range over which x will “run” (here n times, from 0 to n-1).
x will increase by 1 each iteration.
range() can also be given more arguements to define other behaviors, such as starting from a given
value and using a certain step size.
For more info, see here.
for loops with iterables
Certain objects, like lists and strings, are able to be looped over
without using range().
The for loop then becomes something like the following:
for item in list:
do something with item
Every time the loop is run, the next item in the list is being used compared to the previous loop run.
This is generally considered better style-wise than doing:
for i in range(len(list)):
do something with list[i]
which is functionally the same.
Looping over multiple things
One may ask “what if I want to loop over two things at the same time?”, which is a very valid question. A loop for that may be written as follows:
for a, b in zip(list_a, list_b):
do stuff with a and b
This will iterate over list_a and list_b in parallel, stopping when the end of the shortest is reached.
More finely tuned behavior can be achieved, such as running until both list_a and list_b
are exhausted or requiring that the two lists are the same length.
Nested for loops
Nested for loops are like the name implies - a for loop within another for loop.
These are useful if one is working with multidimensional data (i.e., a 2D list)
or combinations of two different variables (say all possible values of \(x^2\) + \(y^2\) for \(x\) and \(y\) between 0 and 10).
While useful, do keep in mind that it tends to be regarded as somewhat inelegant, and in some cases there are ways of avoiding excessive nesting (e.g., vectorization or built-in functions that work on large chunks).
According to the central dogma in molecular biology, genetic information in biological systems flows from DNA to RNA to protein. To make mRNA, a part of a DNA strand is transcribed. The resulting mRNA has nucleotides complementary to the DNA template (note: RNA contains uracil instead of thymine). To produce a protein, ribosomes then use codons (sets of three nucleotides) on mRNA, which are matched with their complementary anticodones on aminoacid-carrying tRNAs.
Let’s go through the process of a) transcription, b) translation, and c) checking for mutations in GFP, everyone’s favourite marker protein.
Pseudocode
For each of the exercises below, start by writing pseudocode on a piece of paper. Only once you’re done with the pseudocode should you start working on Python code in VS Code.
Exercise A: GFP transcription
Write a for loop that will take the string GFP_DNA and convert it to the RNA sequence. Put it into a string called GFP_RNA.
You may assume the input is all capital letters and contains only A, T, C,
and G. Return the GFP_RNA string as capital letters as well.
GFP_DNA = 'ATGAGTAAAGGCGAAGAGCTGTTCACTGGTGTCGTCCCTATTCTGGTGGAACTGGATGGTGATGTCA'\
+ 'ACGGTCATAAGTTTTCCGTGCGTGGCGAGGGTGAAGGTGACGCAACTAATGGTAAACTGACGCTGAAGTTCATC'\
+ 'TGTACTACTGGTAAACTGCCGGTACCTTGGCCGACTCTGGTAACGACGCTGACTTATGGTGTTCAGTGCTTTTC'\
+ 'TCGTTATCCGGACCATATGAAGCGACATGACTTCTTCAAGTCCGCCATGCCGGAAGGCTATGTGCAGGAACGCA'\
+ 'CGATTTCCTTTAAGGATGACGGCACGTACAAAACGCGTGCGGAAGTGAAATTTGAAGGCGATACCCTGGTAAAC'\
+ 'CGCATTGAGCTGAAAGGCATTGACTTTAAAGAAGACGGCAATATCCTGGGCCATAAGCTGGAATACAATTTTAA'\
+ 'CAGCCACAATGTTTACATCACCGCCGATAAACAAAAAAATGGCATTAAAGCGAATTTTAAAATTCGCCACAACG'\
+ 'TGGAGGATGGCAGCGTGCAGCTGGCTGATCACTACCAGCAAAACACTCCAATCGGTGATGGTCCTGTTCTGCTG'\
+ 'CCAGACAATCACTATCTGAGCTACCAAAGCGTTCTGTCTAAAGATCCGAACGAGAAACGCGATCATATGGTTCT'\
+ 'GCTGGAGTTCGTAACCGCAGCGGGCATCACGCATGGTATGGATGAACTGTACAAATAA'
# Your code here
Exercise B: GFP translation
Write a for loop that will take the output string GFP_RNA from Exercise A and
return the protein code GFP_protein (as capitalized one-letter amino acid codes).
You cannot assume that the coding sequence is aligned with the start of your
sequence (i.e., there may be nucleotides in the RNA sequence before the start codon).
Use standard RNA codon table from: https://en.wikipedia.org/wiki/DNA_and_RNA_codon_tables
# Your code here
Exercise C: GFP mutations
Note: the output of exercises A and B is supposed to be the following sequence. If you haven’t solved A and B, continue with this GFP sequence:
MSKGEELFTGVVPILVELDGDVNGHKFSVRGEGEGDATNGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKRHDFFKSAMPEGYVQERTISFKDDGTYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNFNSHNVYITADKQKNGIKANFKIRHNVEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSYQSVLSKDPNEKRDHMVLLEFVTAAGITHGMDELYK
After trying to grow the cells with the protein you just translated, the cells were yellow. It turns out the colleagues who gave us the GFP actually gave us a GFP variant with 4 mutations.
Write a code that gives a list of the amino acid mutations written as XNY, where X is the original amino acid, N is the position of the mutation, and Y is the new amino acid. Example: A mutation of K to G at position 55 is written as K55G.
As input you have:
GFP_protein- your sequence from the previous sectionGFP_reference- a reference sequence
You may assume the sequences are the same length.
GFP_reference = 'MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTL'\
+ 'VTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLV'\
+ 'NRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLAD'\
+ 'HYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK'
# Your code here
Exercise 6.18 ([**] Bacterial population modelling)
Note: this exercise contains an introduction, which is absent from the downloadable exercise (.py) file.
Introduction
while loops
The while loop is another way to repeat an action multiple times and
is especially useful when you do not know how many times you need to loop over a function: while will simply repeat forever while the condition inside is true.
A typical construction is the following:
while error >= 0.0001:
# do some calculation
This calculation will continue until the desired (low enough) error is reached.
An infinite loop can be created in the following way:
while True:
print("This while loop will continue forever")
You should try it - it’s a chance to explore how to stop code from continuing to run in VS Code. Also, accidentally creating infinite loops happens to everyone, so don’t panic if it happens to you :)
A good practice to prevent this from happening excessively is to add a counter in the while loop, which will force
the condition to become false if the loop has been running for too long.
This is an example code with counter i:
some_condition = true
i = 0
while (some_condition and i <= 10000):
print("This while loop will exit after 10000 times")
i += 1 # Increase counter for 1
break and continue
The break and continue statements can be used to further control for and while loops.
break will leave the innermost loop that it is in, skipping the rest of the loop.
This might be useful for things like error cases, or places where you don’t care about the results from some loop.
continue will move to the next iteration of the loop, skipping whatever is below it.
For additional information on control flow, see here.
You are using different types of bacterial populations to potentially use for pharmaceutical synthesis. You want to find out which population is growing the fastest. You have decided that the best way to find out is by doing some simulations of the growth.
Note: break and continue are not necessarily required to solve the two exercises.
Pseudocode
For each of the exercises below, start by writing pseudocode on a piece of paper. Only once you’re done with pseudocode should you start working on Python code in VS Code.
Exercise A: Which strain grows faster?
Write code in which you follow the growth of two bacterial populations, A and B.
We use the following information:
initial_population - initial population of cells
division_t - division time in minutes
If the population of B ever becomes greater than population of A,
set is_B_faster to ‘True’. If not, set is_B_faster to ‘False’.
The maximum time (max_time) you have to simulate is 7 days.
initial_population_A = 120000 # cells
initial_population_B = 241000 # cells
division_t_A = 45 # min
division_t_B = 35 # min
# Your code here
Exercise B: Delayed division start
Do the same as in Exercise A (you can reuse the code you just wrote),
but this time also account for the time before the cells start dividing (start_t),
which is different for the two bacterial populations.
start_t_A = 700 # min
start_t_B = 1200 # min
# Your code here
Exercise C: Race to 1 million cells
Repeat the same idea as in Exercise B, trying to find the fastest-growing strain. This time, however, you are given tuples corresponding to bacterial strains. Each tuple consists of the following: (initial_population, division_t, start_t).
Return the name of the fastest growing strain in best_strain, which is
defined as the quickest strain to reach 1,000,000 cells.
strain_A = (100000, 30, 1000)
strain_B = (120000, 40, 800)
strain_C = (90000, 35, 500)
# Your code here
Exercise 6.19 ([**] Number guess game)
Imagine a number guessing game, where one person chooses a number (let’s say 31) and the other tries to guess it:
Question: “Is it 25?”
Answer: “No, it is higher.”
Question: “Is it 37?
Answer: “No, it is lower.”
Question: “Is it 33?”
Let’s implement this game in Python:
Define a number between 1 and 100.
Ask the user to guess it via the
input()function.After each guess, you have to say whether the answer is correct, or higher or lower than the guess.
The game finishes when the guess is correct.
# Your code here
Exercise 6.20 ([**] CRISPR implementation)
In this exercise you will implement a basic version of CRISPR-Cas9.
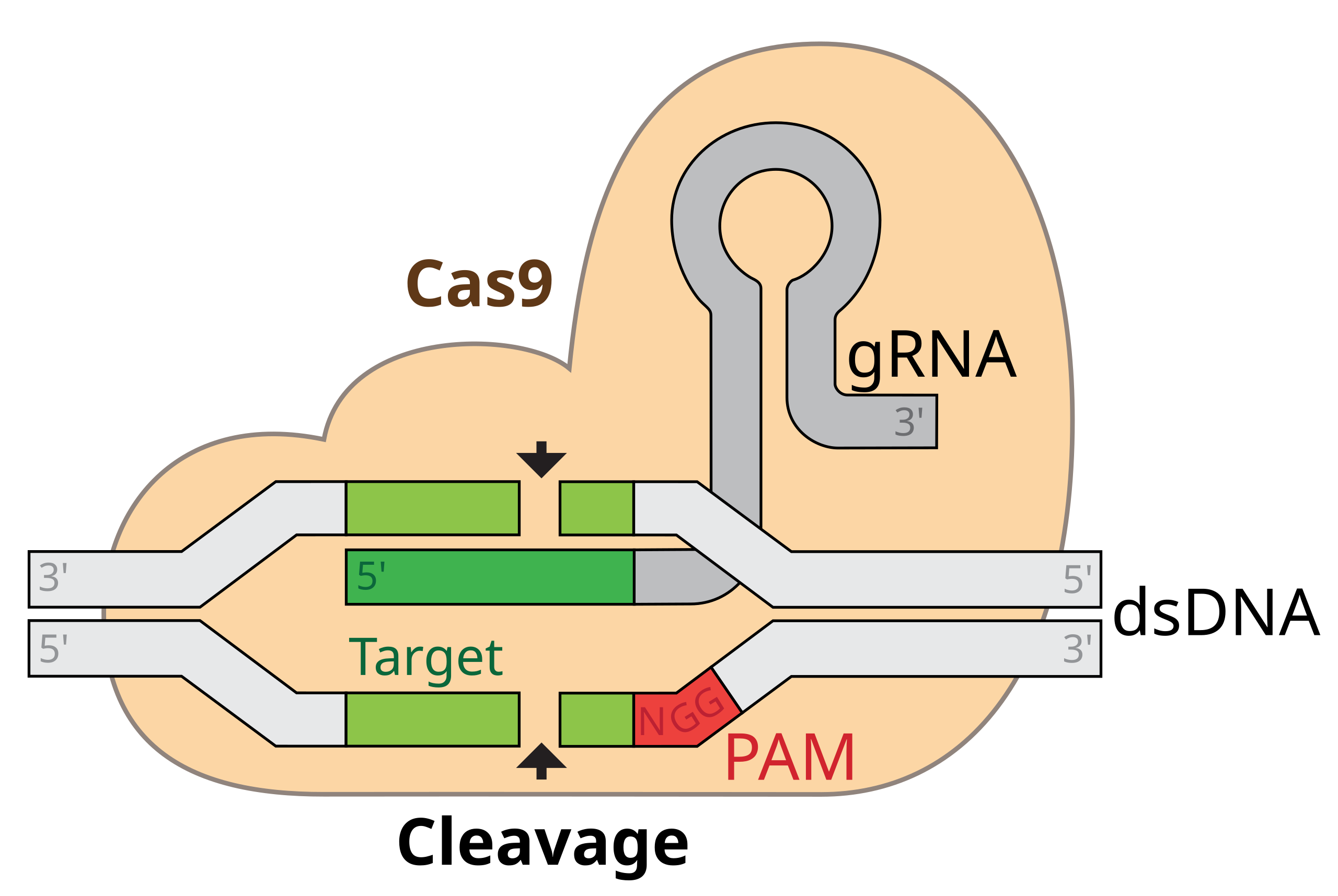
Fig. 6.1 CRISPR-Cas9 (figure source, figure license CC BY-SA 4.0).#
Cas9 protein contains a small piece of RNA called guide RNA (gRNA). This gRNA is used to scan a DNA sequence. If DNA sequence complementary to gRNA is found AND if there is a small protospace adjacent motif (PAM) immediately following the matched sequence, Cas9 will cleave the DNA four nucleotides before the PAM.
Replicate this behavior in Python code.
We will assume that the PAM sequence is NGG, where N stands for any nucleotide (A, C, T, or G). The last two nucleotides are G (guanine).
Inputs:
The DNA sequence to examine
The gRNA sequence to match against the DNA
Both sequences will be given in the 5’ to 3’ direction, the RNA is the complement to the DNA.
Output:
A list containing the sequence after it has been examined by Cas9
E.g., if a sequence was originally ‘ATTGGCC’ and cleaved after ATT, the list looks like this: [‘ATT’, ‘GGCC’].
Hints:
Start by writing pseudocode on a piece of paper. Only once you’re done with pseudocode should you start working on Python code in VS Code.
The input sequence may have more than one cleavage site, or no sites.
The input sequence and guide RNA are reverse complements.
There may be matches with the guide RNA, but not containing the PAM. These should not be cleaved.
You may find it useful to comment out the given DNA and RNA inputs and use your own “toy examples” while working on the code to test whether pieces of your code work.
The
replacefunction may be useful (see more here).
DNA_seq = "CTTTACGACAAGAGGTACGCTTTGTGAGCGCGCTCCGCCGGCCCTCGCGGACCCTTGCTACAAGCTCGACAGGATGTCTCCTCAGCATGTCCTACGTTGGAGGACCGCAACAGCGCCTGGCGGTGCGCACACAGCGGTCGACCGATTTGCGGAGTCCCAGGCCCGGCCGATCCCGTCCCGTACCCTGGTGGACCGTCTAGGGGTCACCTGCCCCTCCGCGATGATTAGATTGCCAGCCCGTAGCGTTTGCGGGTCGCCTACGCGGTCTCTCGGCTGCGCCGGGGATTCGAGCTAGGCCTTTGTAGGCCTGCCGCTGCACAAGGGCCAAAACTCTAACCCTCCCGTTAGCGGGGCGTACCTGGGAGAGGCCATGCCCACGAATGCGCGCCCCGCGGTTGCCTTTTGGGGGCGGGGTCGGCCCTCACGCCGGGGCCCCGTCGGCGCGTCTCTGCGACAAAGGACACCGCGCTGAGCCAAAAGGCTGGCGAGGAACCCGGGAAAAGGGCCCTGGCCCGGTACAGCCCGGGGCGGGGGCTACCCGGGCCTGGAGACCAAATTCGACCACGACCTAGGCCCGACGCGCGGGGCCCATGGCGTAAGCCACAGCAGCTAGAACGCCTTCTCGATAGGCCCACGAATCTGATCGTTGCTATAATAGACGACCTGGCCACCTGGGGAGAATTGAGTTGTTCATGCGCCCCCGGGAAAAGGGCCCTGGCCGTGTACACTCCGTCTAGCCCGAACCGCGAGCCCCACACCCGACCTCGACATGGCCTGGTTGGGTGGGCGGGCGACGGGCGTAGGCCGACCACCGCCGTGGCTGAGACGGCCCCAGAAGAACGTCGCGGGTCAGGCGTTTGAGTGTAACGAGGAACGCTCTTGGTCCACTTGGGGGTTCCCAACAGGTCCAGCGCCCGAGGTGTCGAGATACTGGGGCGCGCGTGGTTGCGGCTACTGGTATATGGATGCACTGGGAGTCCTCTTTGGCCCGGAATGGCTGCACAGCTAACTGGGGGCTCTTGCCCCCATCGGAGCGCGGGAACGCGCCACCGCTGGCA"
gRNA_seq = "GGGCCCUUUUCCCGGG"
# Your code here
Exercise 6.21 ([**] Nanobots)
Simulate the growth of a population of nanobots in a Petri dish. It starts with an initial population of nanobots which have a specific growth rate (e.g., 1.5 per generation). The simulation runs for the select number of generations, and at each generation, the simulation calculates the new population size by multiplying the current population by the growth rate. It then prints the current generation number and the population size. When the simulation is done, print “Nanobot population simulation complete.”.
Implement also the overpopulation condition for the Petri dish: if the population reaches 1 million, all nanobots die. Print a warning at which generation this happened.
If the starting population is zero or below, print a warning.
Try to experiment with different initial parameters.
# Initial population of nanobots
population = 10
# Growth rate (per generation)
growth_rate = 1.5
# Number of generations
generations = 5
# Your code here
Exercise 6.22 ([***] Rock paper scissors (lizard, Spock))
Exercise A
Write an interactive Python implementation of the “Rock paper scissors” game:
Ask the user for their choice (rock, paper, or scissors). Make sure that the user makes a valid choice.
Compare user input with the random computer choice.
Notify the participant whether they won or lost.
Hint: Python’s random package and random.choice() function may come in handy.
# Your code here
Exercise B
Include “lizard” and “Spock” into the game (see the rules here).
# Your code here
Exercise 6.23 ([***] Roman numerals)
Write Python code which transfers decimal numerals into Roman numerals:
I = 1, V = 5, X = 10, L = 50, C = 100, D = 500, M = 1000
In Roman numerals, it is not allowed to have four identical characters consecutively. For example, if we want to convert 41 into Roman numerals, XXXXI is wrong, and XLI is right (think of XLI as 50-10+1).
Exercise A
Use a random user input between 1 and 4000 and automatically rewrite it into Roman numerals. In this part of the exercise, 1249 = MCCXXXXVIIII is allowed, so it is OK to have four consecutive identical characters.
# Your code here
Exercise B
Adapt the code in order to check for four consecutive identical characters and correct the number, such that, e.g., 1209 becomes MCCIX. The largest number is 3999, corresponding to MMMCMXCIX.
# Your code here